News
Juntendo, Kao and PFN Discover Skin Surface Lipids-RNA Patterns Specific to Patients with Parkinson’s Disease
Joint study shows potential for novel diagnostic method using sebum RNAs and machine learning
2021.09.21
TOKYO – September 21, 2021 – The joint research team of Shinji Saiki, associate professor, and Nobutaka Hattori, professor, at Department of Neurology of Juntendo University’s Graduate School of Medicine, Biological Science Research Laboratory of Kao Corporation and Preferred Networks, Inc. (PFN) has discovered that patients with Parkinson’s disease exhibit a specific ribonucleic acid (RNA) pattern in their skin surface lipids (SSL), which are mostly composed of sebum. The research also showed that machine learning with the RNAs in SSL (SSL-RNA) data will potentially be a new non-invasive way to diagnose Parkinson’s disease and facilitate early detection of the disease.
The paper on this research, titled Non-invasive diagnostic tool for Parkinson’s disease by sebum RNA transcriptome profile with machine learning, has been published in Scientific Reports at: https://doi.org/10.1038/s41598-021-98423-9
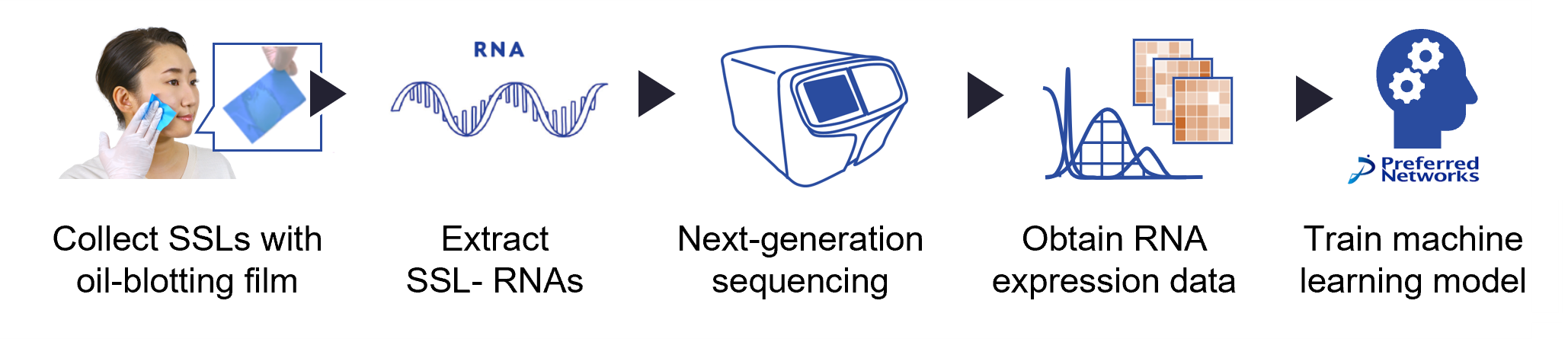 In the joint study, the research team collected facial SSLs with oil-blotting films from Parkinson’s disease patients and healthy controls for comparison. The RNAs were extracted from these SSLs and sequenced using a next-generation sequencer. Analysis of the SSL-RNA expression data identified different patterns between patients with Parkinson’s disease and healthy controls. The research team also trained the machine learning model with the obtained SSL-RNA data, which efficiently distinguished the Parkinson’s disease patients from healthy controls with high accuracy.
In the joint study, the research team collected facial SSLs with oil-blotting films from Parkinson’s disease patients and healthy controls for comparison. The RNAs were extracted from these SSLs and sequenced using a next-generation sequencer. Analysis of the SSL-RNA expression data identified different patterns between patients with Parkinson’s disease and healthy controls. The research team also trained the machine learning model with the obtained SSL-RNA data, which efficiently distinguished the Parkinson’s disease patients from healthy controls with high accuracy.
The research team will continue its research on sebum RNA analysis with machine learning as a non-invasive diagnostic method for similar diseases while validating the findings in Parkinson’s disease.